I got the idea for this assignment while working with bacterial cultures in the lab. The random generation of shapes we did in class kind of reminded me of how bacterial colonies form on agar plates. So, I started by just getting the color of the agar and the bacterial colonies right, similar to the image below.
Fig.: E. coli growing on an agar plate
This is when I also started with experimenting with the randomGaussian() function instead of the regular random() function. This allowed the generation of random positions that were more concentrated towards a central position, basically forming what would be a “3D bell curve”, which usually represents biological growth better than true randomness. And adding translucency to the background gave the effect of some colonies gradually “dying” as new ones “grew”.
However, this alone didn’t look good enough. Off-white colonies on a yellowish background could only look somewhat interesting. That’s when I came across photos of microbial art and also remembered work I did in a class with bacteria that were engineered to be bioluminescent using a Green Fluorescent Protein (GFP) from jellyfish.

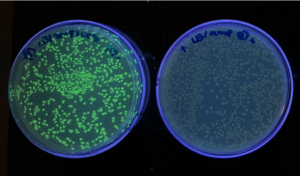 Fig.: (a) Microbial Art from the lab of Prof. Roger Tsien (discoverer of GFP)
Fig.: (a) Microbial Art from the lab of Prof. Roger Tsien (discoverer of GFP)
(b) Plates with GFP-expressing bacteria under black light grown by me
I wanted to incorporate this in the assignment without losing out on the essence of the original appearance of the colonies. Fortunately, this was easy enough with just an if statement that changed the color variables, and a button that toggled the “UV blacklight” between on/off states.
Next, I wanted to add a bit of interactivity to the project, to allow a user to actually create some microbial art instead of having the colonies just randomly generate in a circle in the centre of the viewport. I decided to create discs that were meant to simulate carbohydrate-impregnated paper discs that can be added to agar plates. These special paper discs gradually diffuse the energy source into the agar, instead of the agar already having an energy source in it. In microbiology, this allows for experiments on how different microbes respond to different energy sources. In this project, it allowed me to easily localize the generation of colonies around multiple user-defined points.
I created a class for the discs, which would allow me to quickly generate them when a mouseClicked() event is triggered, while allowing its properties to be used as a basis for the randomGaussian() functions to generate the colonies. This was my first foray into OOP in JavaScript, and the differences from Python took me by surprise.
The next part was arguably the hardest. I wanted to give the user an option to freeze the plate in time and view their resulting artwork. In the lab, we perform a process called fixation that uses formaldehyde or glutaraldehyde to prevent further growth or death of cells, preserving them for as long as we need them, so it would be cool to give that option to the user too. However, including this feature in a way that did not interfere with the UV toggle was challenging. The easiest way to go about doing this is to use the noLoop() function which interrupts the draw() loop. But in that case, the user was stuck with their last UV setting, whether on or off, as that was also part of the draw loop. Instead, I settled on a compromise, using a multidimensional array to capture colony position and size information for every colony of every disk for the last 2 generations. This data was used to recreate the last 2 generations in the “fixed plate”. While this drastically cut down on the number of visible colonies, I was happy with this compromise as it still allowed to toggle between UV on/off states after the plate was fixed.
if (!fixed) {
background(plateColor);
// create arrays to store colony position and size information
prevColonies = colonies;
colonies = [];
for (
let d = 0, colonyX, colonyY, colonyR, colonySpecs;
d < discs.length;
d++
) {
// discs
fill(discColor);
discs[d].display();
// create sub-array to store specific info for each disc
colonies.push([]);
// bacterial colonies
for (let i = 0; i < numColonies; i++) {
// Gaussian random to ensure aggregation towards discs
colonyX = randomGaussian(discs[d].x, spread);
colonyY = randomGaussian(discs[d].y, spread);
colonyR = random(2, 15);
colonySpecs = { x: colonyX, y: colonyY, r: colonyR }; // dictionary containing colony info
colonies[d].push(colonySpecs);
stroke(colonyBorder);
strokeWeight(3);
fill(colonyColor);
ellipse(colonyX, colonyY, colonyR);
}
}
The finishing touches were just adding another button to clear the plate (basically a reset button that resets everything to setup state), tweaking the colors, adding conditions for when the discs could be placed (with warnings for when they couldn’t), and setting up simple counters that would very slowly increase the number of generated colonies and how spread out they were from the disc.
The final result can be seen below. Remember that you cannot interact with the plate when it is being irradiated with UV (that’s a skin cancer risk waiting to happen) or when it’s fixed (well, the bacteria are dead already).
Overall, I am very happy with this assignment. It feels like I found a creative way to present laboratory processes that I work with on a regular basis as a Biology major. If there were any changes I would like to make to it in the future, it would probably be in converting the bacteria to objects as well for easier manipulation. I would also like to include a feature to choose the agar and species growing on it, which can allow experimentation with various interesting colors, such as those produced by some bacteria on blood agar, or by various purple, blue, and other colored bacteria or fungi.
